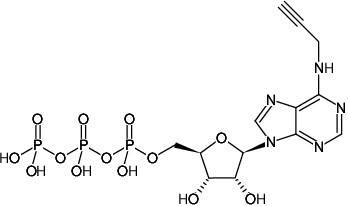
N6-Propargyl-ATP (N6pATP)
N6-Propargyl-adenosine-5'-triphosphate, Sodium salt
| Catálogo Nº | Apresentação | Preço (R$) | Comprar |
|---|---|---|---|
| CLK-NU-001-1 | 1 mg | Sob demanda | Adicionar ao Carrinho |
| CLK-NU-001-5 | 5 mg | Sob demanda | Adicionar ao Carrinho |
For general laboratory use.
Envio: shipped on gel packs
Condições de armazenamento: store at -20 °C
Short term exposure (up to 1 week cumulative) to ambient temperature possible.
Validade: 12 months after date of delivery
Fórmula molecular: C13H18N5O13P3 (free acid)
Peso molecular: 545.23 g/mol (free acid)
Pureza: ≥ 95 % (HPLC)
Forma: solid
Solubilidade: 10 mM Tris-HCl pH 7.5
Propriedades espectroscópicas: λmax 262 nm, ε 18.0 L mmol-1 cm-1 (Tris-HCl pH 7.5)
Formulários:
in vitro AMPylation of proteins[1,2]
in vitro polyadenylation of RNA[3]
The resulting alkyne-functionalized protein[1,2] or RNA[3] can subsequently be processed via Cu(I)-catalyzed (azide-alkyne) click chemistry that offers the choice
- to introduce a Biotin group for subsequent purification tasks (via Azides of Biotin)
- to introduce fluorescent group for subsequent microscopic imaging (via Azides of fluorescent dyes)
- to crosslink the RNA to azide-functionalized biomolecules e.g.proteins
Agonistic ligand, mainly for nucleoside receptor A1
Nucleoside-triphosphates can be converted by different membrane-bound phosphatases into nucleosides acting as nucleoside receptor ligands. In some cases nucleoside phosphates act also directly on nucleoside receptors.
Please note: This compound contains a phosphoramide linkage which is hydrolyzed at pH
Produtos relacionados: Copper (II)-Sulphate (CuSO4), #CLK-MI004 Tris(3-hydroxypropyltriazolylmethyl)amine (THPTA), #CLK-1010 Sodium Ascorbate (Na-Ascorbate), #CLK-MI005
Referências selecionadas:
[1] Grammel et al. (2011) A Chemical Reporter for Protein AMPylation. J. Am. Chem. Soc. 133:17103.
[2] Broncel et al. (2012) A New Chemical Handle for Protein AMPylation at the Host-Pathogen Interface. ChemBioChem 13:183.
[3] Grammel et al. (2012) Chemical Reporter for Monitoring RNA Synthesis and Poly (A) Tail Dynamics. ChemBioChem 13:1112.
[4] Presolski et al. (2011) Copper-Catalyzed Azide-Alkyne Click Chemistry for Bioconjugation. Current Protocols in Chemical Biology 3:153.
[5] Hong et al. (2011) Analysis and Optimization of Copper-Catalyzed Azide-Alkyne Cycloaddition for Bioconjugation. Angew. Chem. Int. Ed. 48:9879.
Sirci et al. (2012) Ligand-, structure- and pharmacophore-based molecular fingerprints: a case study on adenosine A1, A2A, A2B, and A3 receptor antagonists. J. Comput. Aided Mol. Des. 26:1247.
Volonte et al. (2009) Membrane components and purinergic signalling: the purinome, a complex interplay among ligands, degrading enzymes, receptors and transporters. FEBS J. 276:318.
Yegutkin (2008) Nucleotide and nucleoside converting enzymes: Important modulators of purinergic signalling cascade. Biochim. Biophys. Acta 1783:673.
Joshi et al. (2005) Purine derivatives as ligands for A3 adenosine receptors. Current Topics in Medicinal Chemistry 5:1275.
Volpini et al. (2002) N6-Alkyl-2-alkynyl derivatives of adenosine as potent and selective agonists of the human adenosine A3 receptor and starting point for searching A2B ligands. J. Med. Chem. 45 (15):3271.
Hess (2001) Recent advantages in adenosine receptor antagonist research. Expert Opin. Ther. Patents 11 (10):1533.
Jacobson (2001) Probing adenosine and P2 receptors: design of novel purines and nonpurines as selective ligands. Drug Development Res. 52:178.
Jacobson et al. (2001) Ribose modified nucleosides and nucleotides as ligands for purine receptors. Nucleosides, Nucleotides & Nucleic Acids 20 (4):333.
Van Galen et al. (1994) A binding site model and structure-activity relationships for rat A3 adenosine receptor. Molecular Pharmacology 45:1101.
