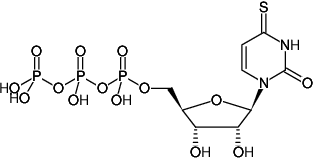
4-Thio-UTP
s4UTP, 4sUTP
4-Thio-uridine-5'-triphosphate, Sodium salt
| Catálogo Nº | Apresentação | Preço (R$) | Comprar |
|---|---|---|---|
| NU-1156S | 10 μl (100 mM) | Sob demanda | Adicionar ao Carrinho |
| NU-1156L | 5 x 10 μl (100 mM) | Sob demanda | Adicionar ao Carrinho |
For general laboratory use.
Envio: shipped on gel packs
Condições de armazenamento: store at -20 °C
Short term exposure (up to 1 week cumulative) to ambient temperature possible.
Validade: 12 months after date of delivery
Fórmula molecular: C9H15N2O14P3S (free acid)
Peso molecular: 500.20 g/mol (free acid)
Pureza: ≥ 95 % (HPLC)
Forma: solution in water
Concentração: 100 mM - 110 mM
pH: 7.5 ±0.5
Propriedades espectroscópicas: λmax 331 nm, ε 16.3 L mmol-1 cm-1 (Tris-HCl pH 7.5)
Formulários:
Potent agonist for P2Y2 and P2Y4 receptors[1,4]
Labeling of transcriptional complex[2]
Influence on elongation and termination events and evoking transcriptional pause[3]
Produtos relacionados:
- HighYield T7 RNA Synthesis Kit, #RNT-101
- HighYield T7 RNA Crosslinking Kit (4-thio-UTP), #RNT-135
Referências selecionadas:
[1] Jacobson et al. (2006) Structure activity and molecular modeling analyses of ribose- and base-modified uridine 5-triphosphate analogues at the human P2Y2 and P2Y4 receptors. Biochemical Pharmacology 71 (4):540.
[2] Khanna et al. (1991) Photoaffinity labelling of the pea chloroplast transcriptional complex by nascent RNA in vitro. Nucleic Acids Res. 19 (18):4849.
[3] Dissinger et al. (1990) Active site labeling of Escherichia coli transcription elongation complexes with 5-[(4-azidophenacyl)thio]uridine 5'-triphosphate. J. Biol. Chem. 265 (13):7662.
[4] Shaver et al. (1997) 4-substituted uridine 5'-triphosphates as agonists of the P2Y2 purinergic receptor. Nucleosides and Nucleotides 16 (7):1099.
[5] Munchel et al. (2011) Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol. Biol. Cell 22 (15):2787.
[6] Zaher et al. (2006) A general RNA-capping ribozyme retains stereochemistry during cap exchange. J. Am. Chem. Soc. 128 (42):13894.
[7] Kwon et al. (2001) DNA sequencing and genotyping by transcriptional synthesis of chain-terminated RNA ladders and MALDI-TOF mass spectrometry. Nucleic Acids Res. 29 (3):e11.
[8] Testa et al. (1999) Thermodynamics of RNA-RNA Duplexes with 2- or 4-Thiouridines: Implications for Antisense Design and Targeting a Group I Intron. Biochemistry 38:16655.
[9] Dontsova et al. (1994) Stem-loop IV of 5S rRNA lies close to the peptidyltransferase center. Proc. Natl. Acad. Sci. USA 91 (10):4125.
[10] Sheng et al. (1993) Active site labeling of HIV-1 reverse transcriptase. Biochemistry 32 (18):4938.
[11] Khanna et al. (1991) Photoaffinity labelling of the pea chloroplast transcriptional complex by nascent RNA in vitro. Nucleic Acids Res. 19 (18):4849.
[12] Tanner et al. (1988) Binding interactions between yeast tRNA ligase and a precursor transfer ribonucleic acid containing two photoreactive uridine analogues. Biochemistry 27 (24):8852.
[13] Bartholomew et al. (1986) RNA contacts subunits IIo and IIc in HeLa RNA polymerase II transcription complexes. J. Biol. Chem. 261 (30):14226.
[14] Eshaghpour et al. (1979) Specific chemical labeling of DNA fragments. Nucleic Acids Res. 7 (6):1485.
