JBS DNA-Shuffling Kit
Random Mutagenesis by DNA Shuffling
| Catálogo Nº | Apresentação | Preço (R$) | Comprar / Observação |
|---|---|---|---|
| PP-103 | 15 reactions | Sob demanda | Adicionar ao Carrinho |
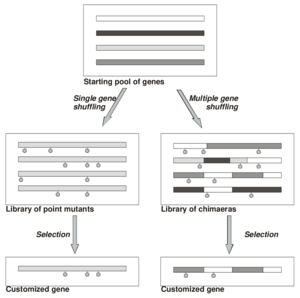
Fig. 1: General types of DNA shuffling
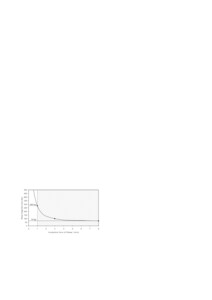
Fig. 2: DNase I digestion of a 1000 bp DNA fragment as a function of incubation time (1 μg DNA + 0.1 unit DNase I at 37°C)
For general laboratory use.
Envio: shipped on gel packs
Condições de armazenamento: store at -20 °C
avoid freeze/thaw cycles
Validade: 12 months
Descrição:
Random Mutagenesis by DNA Shuffling
Developed by Stemmer (1994) DNA shuffling generates libraries by random fragmentation of one gene or a pool of related genes, followed by the reassembly of the fragments in a self-priming PCR reaction. This method allows the recombination of sequences from different, related genes. The overall rate of mutagenesis is approx. 0.7%.
Contente:
DNase I (yellow cap)
0.1 units/μl, 100 μl
Digestion Buffer (blue cap)
10x concentration, 100 μl
DNase Stop Solution (yellow cap)
100 μl
Taq Polymerase (red cap)
5 units/μl, 40 μl
Shuffling Buffer (green cap)
10x concentration, 200 μl
dNTP Mix (white cap)
10 mM each dNTP (dATP, dCTP, dGTP,dTTP), 40 μl
PCR-grade Water (white cap)
3x 1 ml
Random Mutagenesis by DNA Shuffling
DNA shuffling is a powerful technique for directed evolution of proteins in vitro. It generates structural diversity by recombination of gene fragments originating from one or several related genes.
DNA shuffling can be divided into single gene shuffling and multiple gene shuffling (Fig. 1). In single gene shuffling only one gene is digested and subsequently reassembled resulting in point mutations at a rate of approx. 0.7%.
The major application of DNA shuffling in protein evolution is multiple gene shuffling (often referred to as molecular breeding). In this technique several homologous DNA sequences are digested and subsequently reassembled. The result is a library of chimaeric genes containing additional point mutations.
A pivotal step in DNA shuffling is the digest of the gene of interest for production of fragments of appropriate size. Therefore, all reagents in the Kit are optimised to obtain fragments of the desired size within a convenient time frame (Fig. 2).
Usually best results were achieved using fragments with mean sizes of 70-280 bp. Note that a complete DNase I digest results in very short fragments that cannot be amplified by subsequent PCR.
Recommended assay preparation
1. DNase I digest of gene(s) of interest
- For a total volume of 50 μl, use 5 μl of 10x Digestion Buffer in an appropriate vial and add 0.5-2 μg of each starting DNA
- Add PCR-grade Water to a final volume of 50 μl
- Add 0.1 u (1.0 μl) DNase I per μg of starting DNA
- Depending on the desired fragment size incubate at 37°C for up to 8 min (refer to Fig. 2)
- Quench the digest by addition of 5 μl DNase Stop Solution followed by heat inactivation of the DNase I at 75°C for 10 min.
- Isolate fragments of the desired size range by agarose gel electrophoresis and purify using standard procedures.
2. First PCR (without primers)
- For a 50 μl reaction, take 5 μl of the 10x Shuffling Buffer in an appropriate vial and add the purified DNA fragments from step 1 to a final concentration of 10-20 ng/μl
- Add 1 μl of dNTP Mix
- Add 2.5 units (0.5 μl) of Taq Polymerase
- Add PCR-grade Water to a final volume of 50 μl
- Recommended thermocycling conditions:
1. Denaturation: 94°C 90 sec
2. Annealing: 55°C 30 sec
3. Extension: 72°C 30 sec
Number of cycles: 30-45 - Purify PCR product using standard procedures
3. Second PCR (with primers)
- For a 50 μl reaction, take 5 μl of the 10x Shuffling Buffer and add 2 μl of the PCR product from step 2
- Add 1 μl of dNTP Mix
- Add primers to a concentration of 0.8 μM each
- Add 2.5 units (0.5 μl) of Taq Polymerase
- Add PCR-grade Water to a final volume of 50 μl
- Recommended thermocycling conditions:
1. Denaturation: 94°C 30 sec
2. Annealing: 55°C 30 sec
3. Extension: 72°C 30 sec
Number of cycles: 15
Referências selecionadas:
Crameri et al. (1998) DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391:288. Patten et al. (1997) Applications of DNA shuffling to pharmaceuticals and vaccines. Curr. Opin. Biotechnol. 8:724. Stemmer (1994) DNA shuffling by random fragmentation and reassembly. In vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 91:10747. Stemmer (1994) Rapid evolution of a protein in vitro by DNA shuffling. Nature 370:389.
