JBS Error-Prone Kit
Random Mutagenesis by Error-Prone PCR
| Catálogo Nº | Apresentação | Preço (R$) | Comprar / Observação |
|---|---|---|---|
| PP-102 | 15 reactions | Sob demanda | Adicionar ao Carrinho |
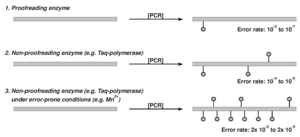
Enhanced mutational rate by error-prone PCR compared to standard PCR reactions.
For general laboratory use.
Envio: shipped on gel packs
Condições de armazenamento: store at -20 °C
avoid freeze/thaw cycles
Validade: 12 months
Descrição:
Random Mutagenesis by Error-Prone PCR
Developed by Caldwell & Joyce (1992) this method introduces mutations in the gene of interest using a PCR reaction under conditions that induce an increased error-rate of the DNA-polymerase. The rate of mutagenesis achieved by error-prone PCR is in the range of 0.6-2.0%.
Contente:
Taq Polymerase (red cap)
5 units/μl, 20 μl
10x Reaction Buffer (blue cap)
10x concentration, 100 μl
10x Error-prone Solution (yellow cap)
10x concentration, 100 μl
dNTP Error-prone Mix (white cap)
unbalanced dNTP ratio (dATP, dCTP, dGTP, dTTP), 40 μl
PCR-grade Water (white cap)
1 ml
Random Mutagenesis by Error-Prone PCR
The standard DNA polymerases used in conventional PCR reactions display error rates that are usually not suitable for directed mutagenesis experiments. For example, proofreading enzymes such as Pfu exhibit error rates in the range from 10-6 to 10-7 whereas non-proofreading enzymes like Taq Polymerase show error rates in the range from 10-4 to 10-5. This rate however, can be significantly enhanced by modifying the following parameters of a PCR-reaction:
- Higher Mg2+concentration of up to 7 mM
- Partial substitution of Mg2+ by Mn2+
- Optimized dNTP concentrations at unbalanced rates
Recommended assay preparation
- For a 50 μl reaction, take 5 μl of 10x Reaction Buffer in a sterile vial and refer to Tab. 1.
- Add 2 μl dNTP Error-prone Mix.
- Add template and appropriate primers. Note that depending on the template the required concentration can be up to 10 times higher compared to standard PCR.
- Add 0.4-1 μl Taq Polymerase as recommended.
- Add PCR-grade Water to a final volume of 45 μl.
- Add 5 μl of 10x Error-prone Solution. Note that the solution should be added last to the reaction mixture to prevent precipitation and must be protected from oxidation (oxidation of Mn2+ to Mn3+ may destroy the polymerase).
Tab. 1: Amounts of components for error-prone PCR conditions (50 μl PCR assay)
| Component | Amount | Final conc. | Cap |
| 10x Reaction Buffer | 5 μl | 1x | blue |
| dNTP Error-prone Mix | 2 μl | unbalanced ratio | white |
| Primers | 20-100 pmol | ||
| Template | 3-100 fmol / 2 - 50 ng | ||
| Taq Polymerase | 0.4-1 μl | 2-5 units | red |
| PCR-grade Water | Fill up to 45 μl | white | |
| 10x Error-prone Solution | 5 μl | 1x | yellow |
Recommended thermocycling conditions
| Denaturation | 94°C | 30 sec |
| Annealing1) | approx. 45-68°C | 30 sec |
| Extension2) | 72°C | 1 min |
Number of cycles: 30
1) The annealing temperature depends on the melting temperature of the primers.
2) The elongation time depends on the length of the fragments to be amplified. A time of 1 min per kbp is recommended.
For optimal specificity and amplification an individual optimization of the recommended parameters may be necessary for each new template DNA and/or primer pair.
Referências selecionadas:
Kim et al. (2001) Improvement of tagatose conversion rate by genetic evolution of thermostable galactose isomerase. Biotechnol. Appl. Biochem. 34:99.
Daugherty et al. (2000) Quantitative analysis of the effect of the mutation frequency on the affinity maturation of single chain Fv antibodies. PNAS 97:2029.
Wan et al. (1998) In vitro evolution of horse heart myoglobin to increase peroxidase activity. Proc. Natl. Acad. Sci. USA 95:12825.
Cline et al. (1996) PCR fidelity of Pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Res. 24:3546.
Vartanian et al. (1996) Hypermutagenic PCR involving all four transitions and a sizeable proportion of transversions. Nucleic Acids Res. 24:2627.
Cadwell et al. (1992) Randomization of genes by PCR mutagenesis. PCR Meth. Appl. 2:28.
Kunkel (1992) DNA replication fidelity. J. Biol. Chem. 287:18251.
