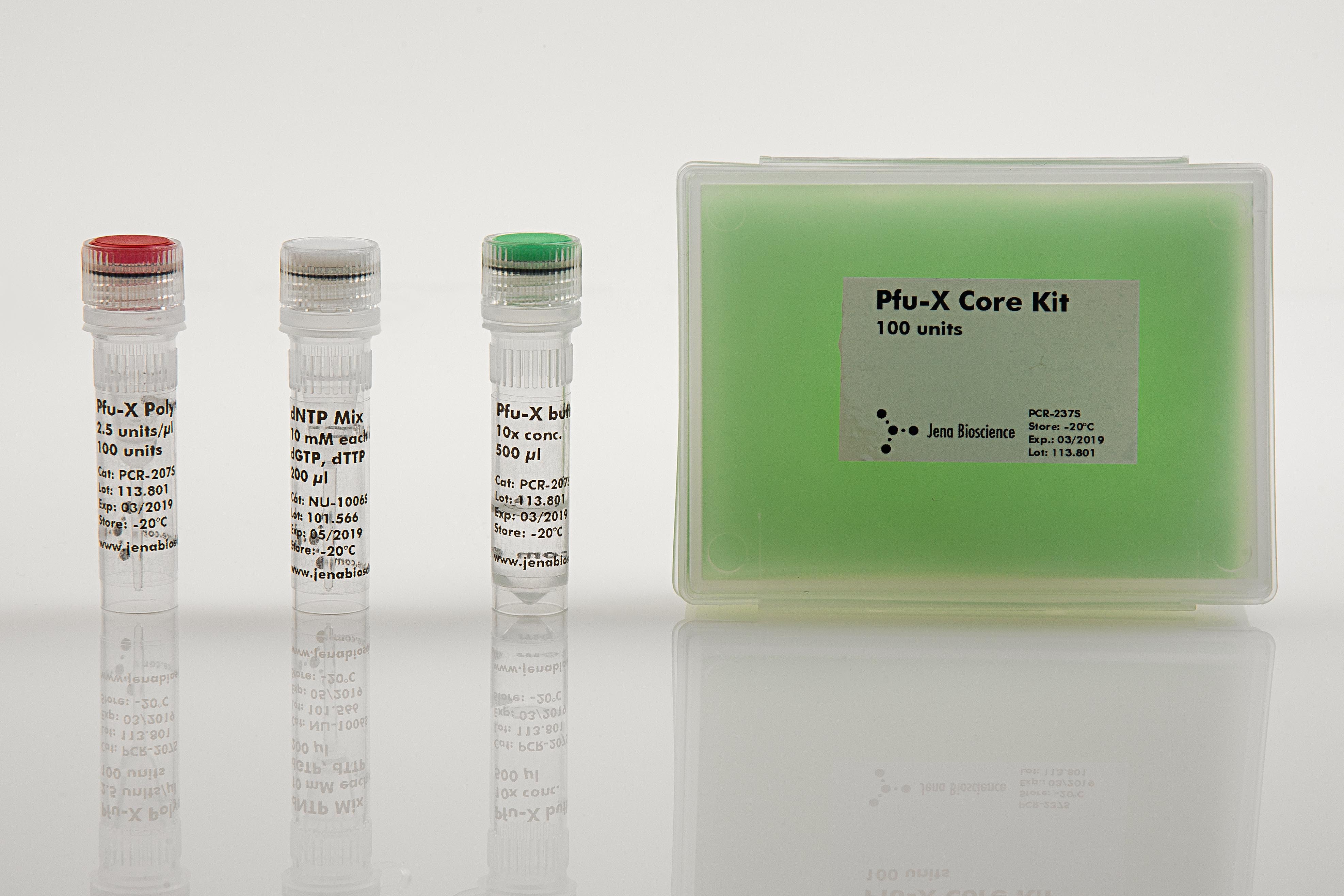
Pfu-X Core Kit
Kit for high accuracy PCR and Ligase-free cloning
| Catálogo Nº | Apresentação | Preço (R$) | Comprar |
|---|---|---|---|
| PCR-237S | 100 units | Sob demanda | Adicionar ao Carrinho |
| PCR-237L | 500 units | Sob demanda | Adicionar ao Carrinho |
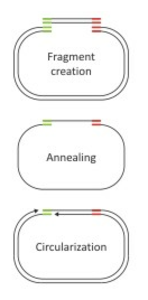
Fig. 1: Functionality of Ligase-free Cloning
For general laboratory use.
Definição de unidade: One unit is defined as the amount of the enzyme required to catalyze the incorporation of 10 nmol of dNTP into an acid-insoluble form in 30 minutes at 74 °C.
Envio: shipped on gel packs
Condições de armazenamento: store at -20 °C
avoid freeze/thaw cycles
Validade: 12 months
Forma: liquid
Concentração: 2.5 units/μl
Descrição:
Pfu-X Core Kit contains all reagents required for PCR (except template and primer) in one box combining simple handling with high flexibility. The premium quality polymerase, ultrapure dNTPs and the optimized complete reaction buffer ensure superior amplification results.Pfu-X Polymerase is the ideal choice for applications where the efficient amplification of DNA with highest fidelity is required.The enzyme is a genetically engineered Pfu DNA polymerase, showing a 2-fold higher accuracy and an increased processivity, resulting in shorter elongation times.The enzyme catalyzes the polymerization of nucleotides into duplex DNA in 5'→3' direction but does not possess a 5'→3' exonuclease replacement activity. Its inherent 3'→5' exonuclease proofreading activity results in a greatly increased fidelity of DNA synthesis compared to Taq polymerase. Pfu-X Polymerase-generated PCR fragments are blunt-ended.The enzyme is highly purified and free of bacterial DNA.Fidelity of the enzyme:Pfu-X Polymerase is characterized by a 50-fold higher fidelity compared to Taq polymerase and a 2-fold higher fidelity compared to standard Pfu polymerase.ERPfu-X Polymerase = 0.25 x 10-6The error rate (ER) of a PCR reaction is calculated using the equation ER = MF/(bp x d), where MF is the mutation frequency, bp is the number of base pairs of the fragment and d is the number of doublings(2d = amount of product / amount of template).
Contente:
Component||PCR-237S||PCR-237L
Pfu-X Polymerase2.5 units/μlin storage buffer*red cap||40 μl100 units||200 μl500 units
dNTP Mix10 mM eachdATP, dCTP,dGTP, dTTPwhite cap||100 μl||500 μl
Pfu-X Buffer10x conc.green cap||500 μl||2 x 1.2 ml
PCR-grade Waterwhite cap||2 x 1,2 ml||2 x 6 ml
* (50 % Glycerol, 50 mM Tris-HCl pH 8.0, 0.1 mM EDTA, 1 mM DTT 0.1 % Tween 20, 0.1 % Nonidet P-40)
Recommended 50 μl PCR assay:5 μl||10x Pfu-X Buffer||green cap
1 μl||dNTP Mix||white cap
0.4 μM||each Primer||-
1 - 100 ng||template DNA||-
0.5 μl(1.25 units)||Pfu-X Pol||red cap
Fill up to 50 μl||PCR-grade water||-
Please note that it is essential to add the polymerase as last component.
Recommended cycling conditions:
Three-step standard protocolinitialdenaturation||95 °C||2 min||1x
denaturation||95 °C||20 sec||25-30x
annealing1)||50 - 68 °C||30 sec||25-30x
elongation2)||68 °C||1 min/kb||25-30x
finalelongation||68 °C||1 min/kb||1x
Two-step protocol for amplification of longer fragments (>3 kb)
Please note that for performing two-step cycling a sufficiently high primer Tm is necessary. If Tm of primers is below 65 °C or two-step PCR does not yield a sufficient product quality the three-step cycling protocol is recommended.initialdenaturation||95 °C||2 min||1x
denaturation||95 °C||20 sec||25-30x
annealing/elongation1,2)||68 °C||30 sec/kb||25-30x
finalelongation||68 °C||30 sec/kb||1x1)The annealing temperature depends on the melting temperature of the primers used.
2)The elongation time depends on the length of the fragments to be amplified. A time of 1 min/kb is recommended.
For optimal specificity and amplification an individual optimization of the recommended parameters may be necessary for each new template DNA and/or primer pair.
Ligase-free Cloning
Ligase-free Cloning is based on a cloning technique invented by Quan and Tian in 2009. It offers a number of advantages over conventional cloning methods. The system:
works with any vector that can be linearizedallows efficient cloning even into blunt end vectorsallows directed cloning into single-cut vectorsallows fast and easy preparation of vector and insert with no or only few purification stepsdoes not require to dephosphorylate the vectorallows the use of any restriction enzyme that linearizes the vector even if its recognition site(s) are present in the insertdoes not need a ligation stepdoes not add additional sequences to the plasmid or the insert
Principle (see Fig. 1)
Ligase-free Cloning is based on generation of inserts with homologous ends to the linearized vector.
In a circularization reaction, vector and insert anneal due to their homologous ends.
Using a specially selected DNA polymerase, the resulting single-stranded plasmids are recircularized.
These plasmids can directly be used for transformation. They still have two nicks each, which will be repaired by E. coli's endogenous DNA repair system and thus do not have to be ligated in vitro.
Supplements (to be provided by user)
Produtos relacionados: Ready-to-Use Mixes / direct gel loading Ready-to-Use Mixes Thermophilic Polymerases Deoxynucleotides (dNTPs) Supplements Primers and Oligonucleotides DNA Ladders
Referências selecionadas:
Quan et al. (2009) Circular polymerase extension cloning of complex gene libraries and pathways. PLoS One. 4:e6441.
