2-Fluoro-ATP
(2F-ATP)
2-Fluoro-adenosine-5'-triphosphate, Sodium salt
| Catálogo Nº | Apresentação | Preço (R$) | Comprar |
|---|---|---|---|
| NU-145L | 5 x 10 μl (100 mM) | Sob demanda | Adicionar ao Carrinho |
| NU-145S | 10 μl (100 mM) | Sob demanda | Adicionar ao Carrinho |
For general laboratory use.
Envio: shipped on gel packs
Condições de armazenamento: store at -20 °C
Short term exposure (up to 1 week cumulative) to ambient temperature possible.
Validade: 12 months after date of delivery
Fórmula molecular: C10H15N5O13P3F (free acid)
Peso molecular: 525.17 g/mol (free acid)
CAS#: 1492-62-2 (free acid)
Pureza: ≥ 95 % (HPLC)
Forma: solution in water
Concentração: 100 mM - 110 mM
pH: 7.5 ±0.5
Propriedades espectroscópicas: λabs 261 nm, ε 14.3 L mmol-1 cm-1 (Tris-HCl pH 7.5)
Descrição:
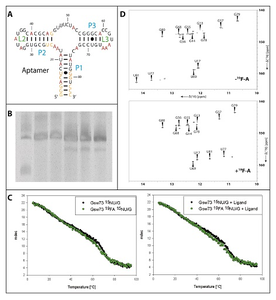
Figure 1A: Schematic representation of the secondary structure of the Gswapt. The stems are named in blue and the loops are named in green. All fluorinated bases are labeled in red.
Figure 1B: 10% PAA-Gel of the transcription samples containing from left to right: low range ssRNA-marker, transcription with unlabeled dNTPs, 15N-UTP, 15N-GTP, 2F-ATP, 15N-UTP-GTP + 2F-ATP.
Figure 1C: CD-melting curve of the Gswapt in the ligand bound state. The black curve represents the unlabeled RNA and the green curve represents the 19F-labeled RNA. The melting curves were recorded in the absence (left) and presence (right) of the ligand.
Figure 1D: 1H-15N-HSQC of the Gswapt. The spectrum of the unlabeled RNA is shown on the top and the spectrum of the 19F-labeled RNA is shown at the bottom.
RNA Structure Determination by NMR with 2F-ATP
Application Note
By courtesy of
Florian Sochor, sochor@nmr.uni-frankfurt.de
and
Harald Schwalbe, schwalbe@nmr.uni-frankfurt.de
Goethe-Universität Frankfurt am Main,
Max-von-Laue-Straße 7, D-60438 Frankfurt am Main
Multidimensional NMR studies of RNAs are based on the correlation between magnetic properties of protons and of other NMR-active atoms. In nucleic acids these atoms are usually 13C, 15N and 31P. Whereas 31P is at 100% natural abundance, the other NMR-active, non-radioactive isotopes have low natural abundance. The methodic challenges are to artificially enrich or introduce NMR active isotopes of 13C and 15N into the molecules without affecting their overall structural integrity. Besides the atoms present in RNA, also introduction of different NMR reporter signals is of considerable interest in NMR studies of large RNAs.
In this application note we substituted the hydrogen at positions 2 in the base of adenosine (Ade-2H) with 19F, which has a comparable gyromagnetic ratio as 1H but exhibits larger chemical shift dispersion. The Ade-2H position was chosen as the attached nucleus is spatially close enough to the base pairing but does not show J-coupling to other protons.
We introduce 19F labelling for the 73 nucleotides comprising RNA aptamer region of the guanine sensing riboswitch from Bacillus subtilis (Gswapt, shown in Fig 1A). Due to the inherent low sensitivity, samples of RNA for NMR spectroscopic studies require high concentration of the solute, in this case of the 19F labeled RNA. Therefore it was synthesized in large scale (0.15 mg) by in vitro transcription with 2F-ATP complementing ATP. The composition of the transcription mix is shown in Table 1. The concentrations of Mg(OAc)2, DNA-template, nucleotides and YIPP were optimized for highest transcription efficiency. Several in vitro transcriptions with different nucleotide composition were performed overnight at 37 °C (Figure 1B). The yield of 19F labeled Gswapt synthesized within a transcription reaction containing also 15N-GTP and 15N-UTP was compared with the yield in a transcription reaction containing solely 15N-GTP and 15N-UTP as isotope labeled compounds. All NTP compositions led to a single 73 nt RNA product. The transcription efficiency, measured by HPLC, is 1.9% (±0.4%) for all transcriptions including 2F-ATP compared to 2.1% (±0.5%) for the transcriptions with unlabeled or 15N-labeled nucleotides only.
Table 1: Transcription parameters
Fixed parameters||
Tris HCl pH 8.1||40 mM
DTT||4 mM
Spermidine||1 mM
BSA||50 μg/ml
Triton X-100||0.01 %
Polyethylenglycol||80 mg/ml
T7 RNA-Polymerase||70 μg/ml
Variable parameters||
Mg(OAc)2||27.25 mM
DNA template||67 nM
ATP/2F-ATP||2 mM
CTP||2.24 mM
GTP||2.36 mM
UTP||2 mM
YIPP||0.002 U/μl
RNA stability was investigated by CD-spectroscopy (Figure 1C). In denaturation experiments, the melting curves of the 19F-labeled and the unlabeled RNA show an identical progression. Without ligand, the ΔTm = Gswapt - 19F-Gswapt = 4 K. In the ligand bound state, the ΔTm = 2 K. Furthermore, 1H-15N-HSQCs were recorded with the 19F-labeled and the unlabeled RNA (Fig.2D). The signals corresponding to protons of G-C and G-U-base pairing are identical for both RNAs. In contrast, the signals of the A-U base pairing protons display a highfield shift of = -2.6 ppm.
Produtos relacionados: HighYield T7 P&L RNA NMR Kit (2F-ATP), #RNT-203
Referências selecionadas:
Sochor et al. (2016) S(19)F-labeling of the adenine H2-site to study large RNAs by NMR spectroscopy. J. Biomol. NMR 64 (1):63.
Stockman (2008) 2-Fluoro-ATP as a versatile tool for 19F NMR-based activity screening. J. Am. Chem. Soc. 130 (18):5870.
Scott et al. (2004) Enzymatic synthesis and 19F NMR studies of 2-fluoroadenine-substituted RNA. J. Am. Chem. Soc. 126 (38):11776.
